How Does miR-122 Micro-manage HCV Infection?
Hepatitis C virus (HCV) is a global health problem that affects approximately 71 million people worldwide, including over 268,000 Canadians. HCV-infected individuals typically develop a persistent infection that leads to chronic hepatitis, cirrhosis, steatosis (fatty liver), and liver cancer. To establish an infection, HCV commandeers many different host factors in our cells. In healthy cells, microRNAs typically restrict gene expression by binding to the 3' untranslated (UTR) of target mRNAs. During HCV infection, a highly abundant, liver-specific human microRNA, called miR-122, binds to the 5' end of the HCV genome and promotes viral RNA accumulation; however, the detailed mechanism(s) have remained unclear. Our work demonstrated that miR-122 has extensive interactions with the HCV genome beyond the “seed” sequences, creating a 3´ overhang. The 3´ overhang suggested a role for miR-122 in protection of the HCV 5´ terminus from attack by nucleases or cellular sensors of RNA (Machlin et al PNAS 2011 *Cozzarelli Prize). More recently, we have demonstrated that miR-122 protects the HCV genome from cellular pyrophosphatases and subsequent exoribonuclease-mediated decay (Amador-Cañizares et al NAR 2018). In a related study, we also found that GB virus B is also subject to this regulation, revealing a novel mechanism for RNA accumulation in hepaciviruses (Sagan et al J Virol 2013; Sarnow & Sagan Ann Rev Virol 2016; Bernier & Sagan Viruses 2018).
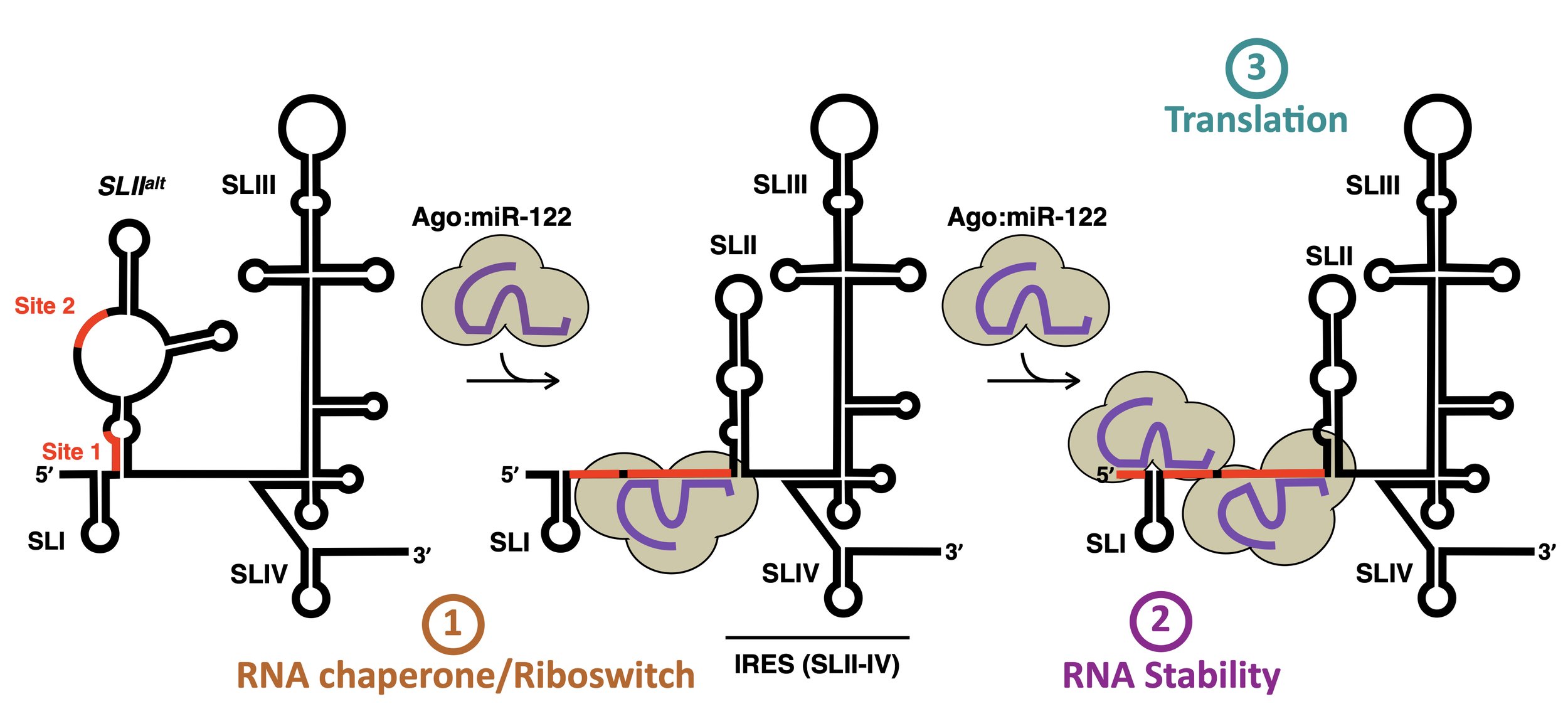
Through three recent studies (Amador-Cañizares et al NAR 2018; Chahal et al NAR 2019; Chahal et al PNAS 2021), we provided evidence that miR-122 has at least 3 roles in the HCV life cycle. Firstly, it acts as an RNA chaperone or riboswitch to re-fold the 5´ UTR and allow the viral internal ribosomal entry site (IRES) to form. Secondly, it protects the 5´ terminus of the HCV genome from 5´ decay. And thirdly, through the associated Argonaute (Ago) protein, it forms direct contacts with the IRES that promote translation. We hypothesize that miR-122 forms a distinct complex with host and/or viral proteins that together mediate viral RNA accumulation and serve as the switch from translation to replication in the HCV life cycle.
We use a combination of point mutations, mutant microRNAs, and reporter viral RNAs to evaluate the contributions of miR-122 in the HCV life cycle. Additionally, we use infectious virus, and assays for viral RNA accumulation (RT-qPCR, Northern blot) as well as focus-forming unit (FFU) assays to assess virion production.
Our recent results suggest that the riboswitch appears to have a minimal contribution in isolation, while genome stability and translational promotion have similar contributions during the establishment phase of infection (Rheault et al. NAR 2023). However, during the maintenance phase, translational promotion becomes the dominant role. Additionally, we found that the alternative conformation of the 5’ UTR, SLIIalt, is important for efficient virion production, suggesting that miR-122 also controls the balance between viral RNAs in the translating/replicating pool and those engaged in virion assembly. More recent preliminary data suggests that miR-122 may mediate the switch from translation to replication and replication organelle biogenesis, perhaps by taking advantage of the canonical RNA silencing pathway afterall…
Open questions
How does HCV escape canonical miRNA-silencing (or does it…)?
What host and/or viral proteins and RNA molecules are part of the miRNA-enhancing complex at the 5’ end of the viral RNA?
Does miR-122 help mediate the switch from translation to replication and/or replication organelle biogenesis?
How are replication organelles formed?
Do other viruses use microRNAs in a similar manner?
Significance
This line of research has already revealed a new mechanism for miRNA-mediated viral RNA accumulation, and we anticipate that this research will reveal new paradigms for miRNA-mediated gene regulation, provide insights into novel mechanisms of resistance to miRNA-based therapies, and will reveal host-virus interactions that may be applicable to other important human and veterinary pathogens.
Check out some of our work on the topic:
Rheault M, Cousineau SE, Fox DR, Abram QH, and Sagan SM. Elucidating the distinct contributions of miR-122 in the HCV life cycle reveals insights into virion assembly (2023). Nucleic Acids Res. 51(5):2447-2463.
Cousineau SE, Rheault M, and Sagan SM. Poly(rC)-binding protein 1 limits hepatitis C virus virion assembly and secretion. (2022) Viruses 14(2):291.
Chahal J, Gebert LFR, Camargo C, MacRae IJ, and Sagan SM*.miR-122-based Therapies Select for Three Distinct Resistance Mechanisms Based on Alterations in RNA Structure. (2021) PNAS USA. Aug 17; 118(33):e2103671118.
Chahal J, Gebert LFR, Gan HH, Camacho E, Gunsalus KC, MacRae IL, and Sagan SM. miR-122 and Ago interactions with the HCV genome alter the structure of the viral 5' terminus. (2019) Nucleic Acids Res. Apr 3. In press.
Bernier A and Sagan SM. Beyond sites 1 and 2, miR-122 target sites in the HCV genome have negligible contributions to HCV RNA accumulation in cell culture. (2019) J Gen Virol. 100(2): 217-226.
Bernier A and Sagan SM. The diverse roles of microRNAs at the host-virus interface. (2018) Viruses. 10(8):E440
Amador-Cañizares Y, Bernier A, Wilson JA, Sagan SM. miR-122 does not impact recognition of the HCV genome by innate sensors of RNA but rather protects the 5’ end from the cellular pyrophosphatases, DOM3Z and DUSP11. (2018) Nucleic Acids Research. 46(10): 5139-5158.
Sarnow P and Sagan SM. Unraveling the Mysterious Interactions Between Hepatitis C Virus RNA and Liver-specific MicroRNA miR-122. Annual Review of Virology (2016) 3(1):309-332.
Sagan SM, Chahal J, and Sarnow P. cis-acting RNA elements in the Hepatitis C virus RNA genome. Virus Res (2015) S1068-1702(14):00543-7.
Wilson JA and Sagan SM. Hepatitis C virus and human miR-122: insights from the bench to the clinic. Curr Opin Virol (2014) 7C:11-18.